How do organisms use the genetic information encoded in their DNA to reliably shape themselves? The laboratory is interested in understanding morphogenesis in plants, which unlike animals, form organs throughout their whole life. We study the formation of lateral roots that determine the plant’s capacity to forage its environment for nutrients and stabilize its anchoring. To understand how these new roots are formed, we combine molecular genetics with cell biology, imaging and quantitative analysis.
Our research
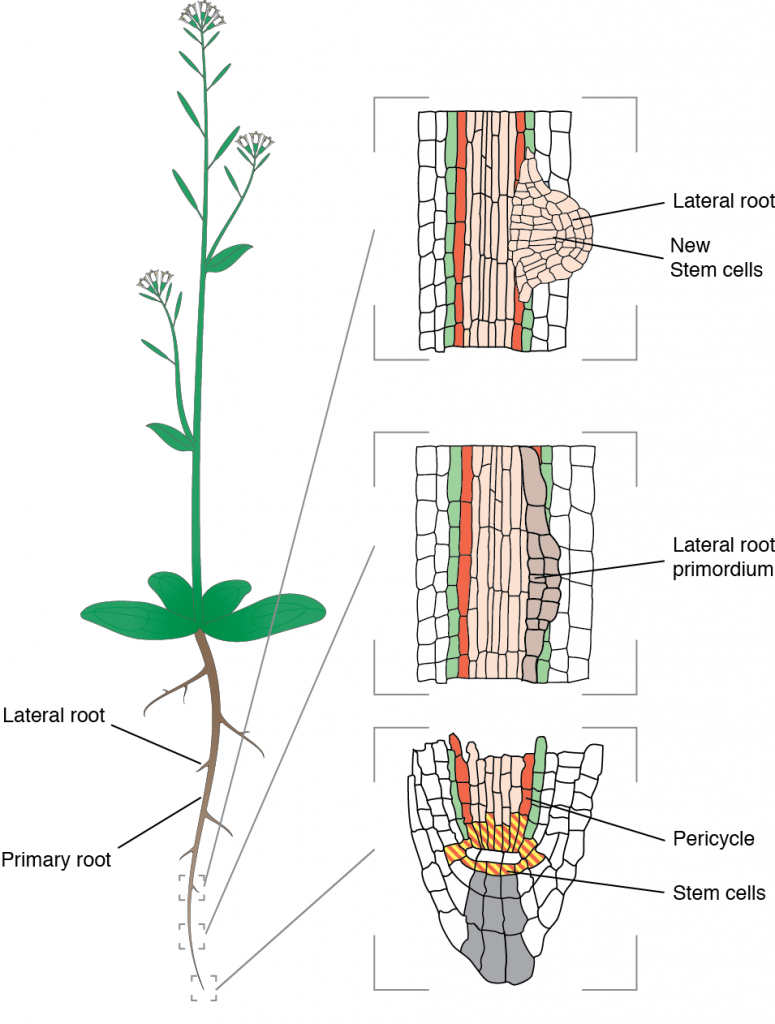
Our model is Arabidopsis thaliana which form lateral roots from founder cells located deep in the primary root. The process happens in the pericycle and debuts with the asymmetric expansion of two abutting founder cells that share a common interface.
These founder cells expand more along this interface and this prefigures the future dome-shaped lateral root primordium that grows out of the parental root. Accompanying the asymmetric growth, both nuclei migrate toward the common cell wall and the cells divide asymmetrically. Failure to execute these steps leads to aberrantly shaped lateral root or complete arrest.
Lateral root formation is a self-organizing process during which founder cells invariably undergo a first asymmetric anticlinal division. In the resulting lateral root initiation site, small daughter cells are positioned next to each other in the center and are flanked by larger ones. We showed that radial expansion of the founder cells and spatial accommodation by the overlying endodermis are essential for the formation of a lateral root and the first asymmetric cell divisions to occur. We have made key discoveries about the role of the cell wall and the cytoskeleton in this process. We have also co-developed innovative methods to analyse the shape of cells.
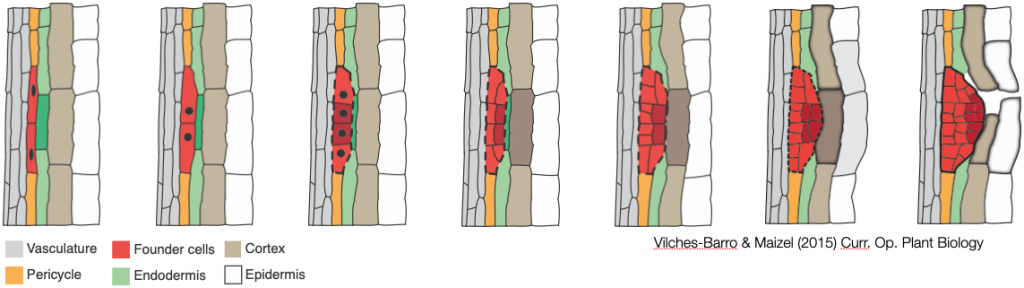
Lateral root formation is a self-organizing process during which founder cells invariably undergo a first asymmetric anticlinal division. In the resulting lateral root initiation site, small daughter cells are positioned next to each other in the center and are flanked by larger ones. We showed that radial expansion of the founder cells and spatial accommodation by the overlying endodermis are essential for the formation of a lateral root and the first asymmetric cell divisions to occur. We have made key discoveries about the role of the cell wall and the cytoskeleton in this process. We have also co-developed innovative methods to analyse the shape of cells.
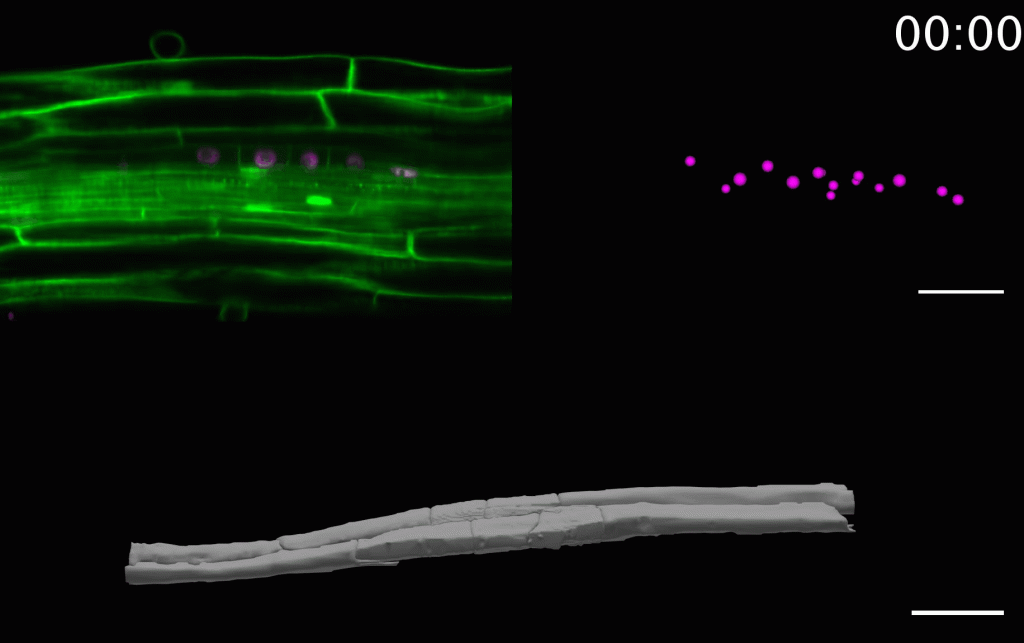
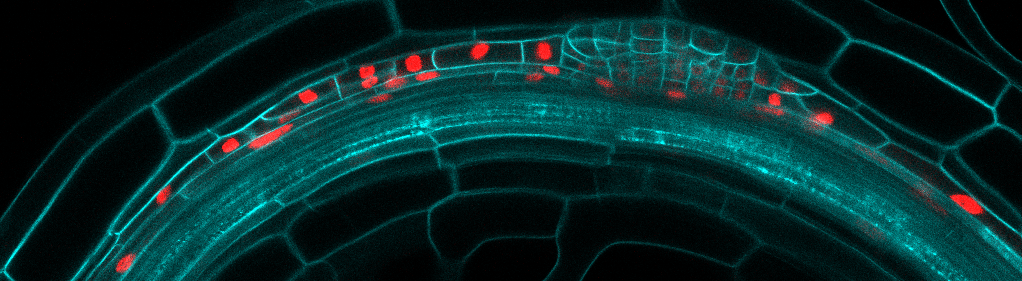
The laboratory is combining precise manipulation of cell properties at cell and tissue scale with live imaging and quantitative analysis to understand the mechanisms responsible for the proper execution of the lateral root initiation program. We have developed several techniques to study and analyze the morphogenesis of lateral roots as it happens.
Impact

Plants harvest energy from sunlight in their leaves to produce sugar. To support their growth they must also forage for water and mineral nutrients with their roots.
Designing better roots represents a under-explored way to increase yield, reduce fertilizer use and pollution, promote soil health and ultimately help people.
How to optimize root systems? It is not a easy question. Building more roots represent a construction cost as well as maintenance cost. Optimizing root systems requires a basic understanding of the cellular mechanism that control how new roots are formed.
Our work by unraveling the basic concepts and mechanisms that underpin the formation of roots, will pave the way for the rational design of better root systems in crops.
Funding
The Maizel lab started as a independent group in the CellNetworks Cluster of Excellence. The laboratory is part of the collaborative research centres SFB1101 on specificity on plant processes, the FOR2581 on plant Morphodynamics and the HBIGS graduate school. Our work is supported by funds from a variety of public sources.
